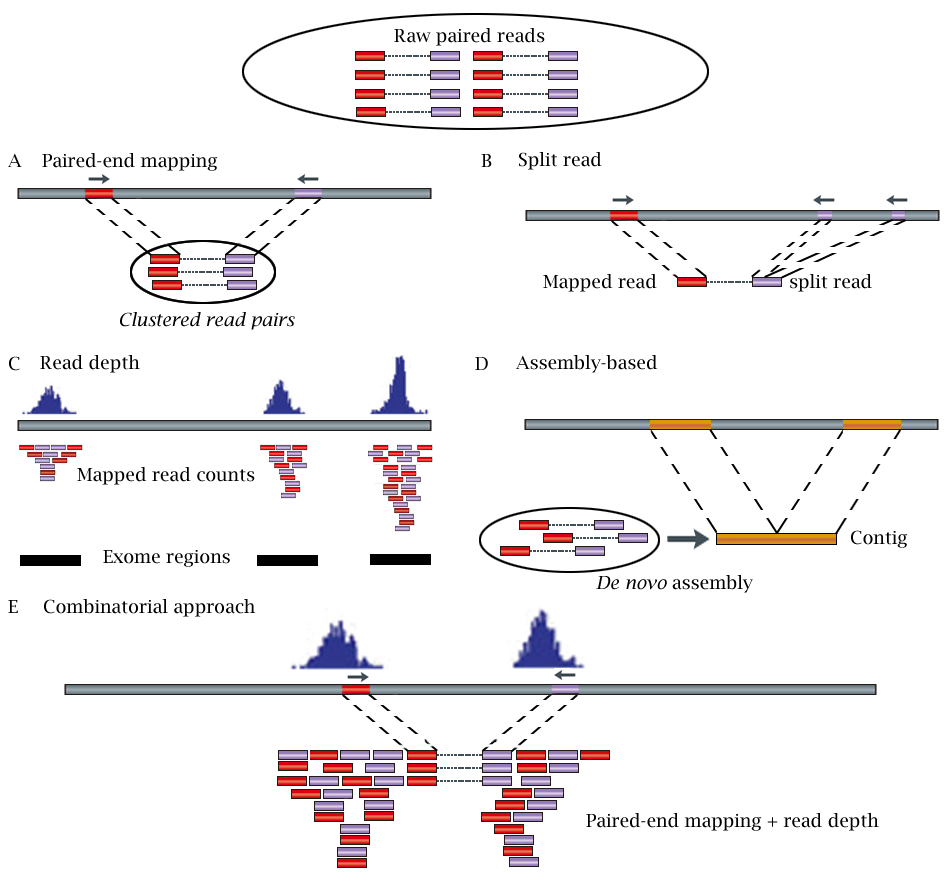
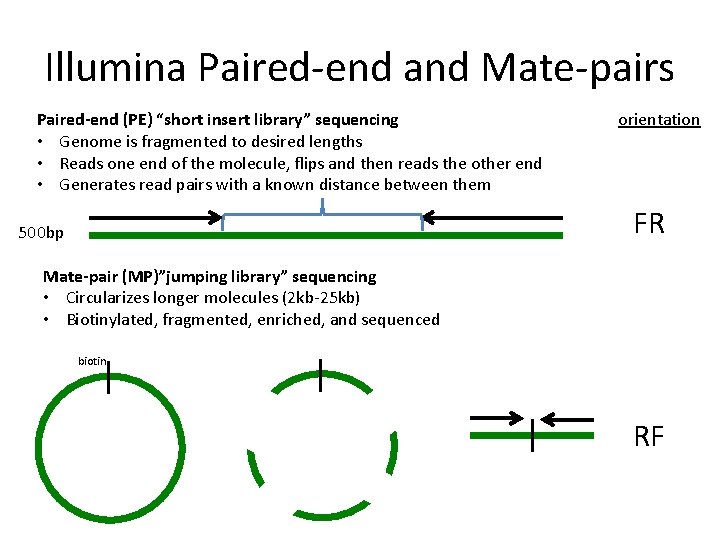
paired end sequencing vs mate pair
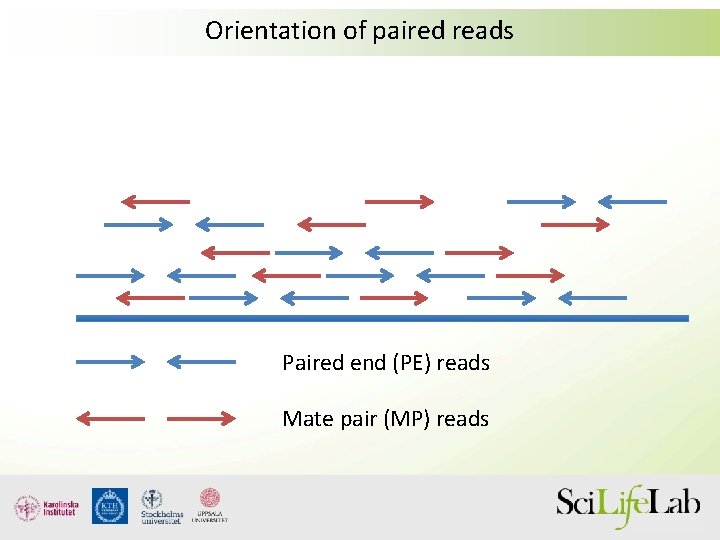
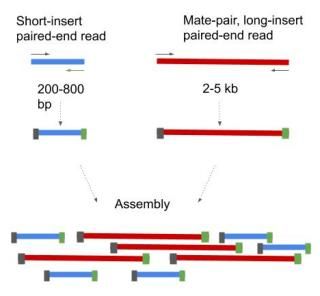
Its a smart technique that allows you to obtain paired-end reads with long insertsFirst DNA is fragmented and fragments of a desired length around 2-5 kbs a. This is different from FR because it means the reverse read aligned at a lower base pair position than the forward read and thus that they are pointing away from another.

Illumina Sequencing Illumina Sequencing By Synthesis 1010genome Quality Ngs Bioinformatics Data Analysis Services
They are not two different methods.
. Requires the same amount. Paired end sequencing reffers to sequrncing of fragments from both ends this is in contrast to single end sequemcing where sequencing is done from one end. Simple workflow allows generation of unique ranges of insert sizes.
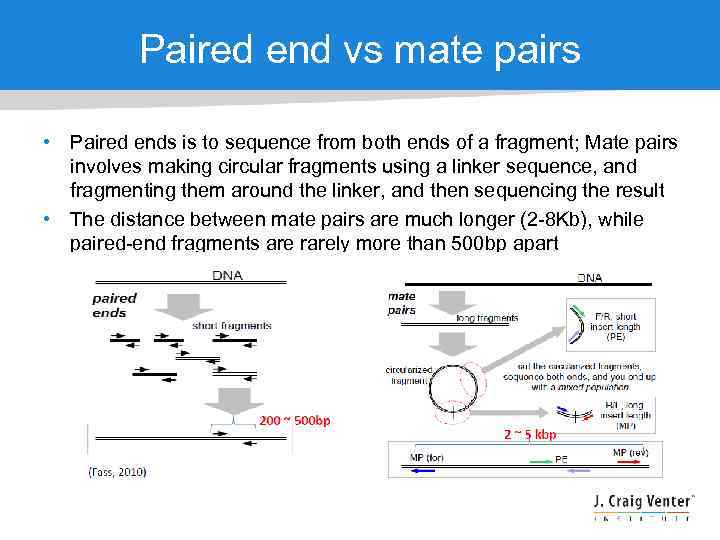
The larger inserts mate pairs can pair reads across greater distances. They are all very different in separate regards but they all refer to different wet-lab and sequencing protocolstechnologies. The decision to use mate-pair vs.
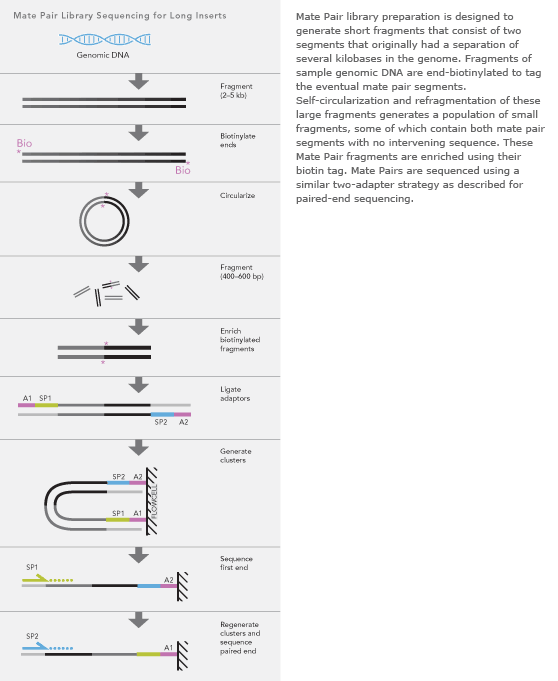
Introduction to Mate Pair Sequencing. The figure shows the. The figure shows the workflow for mate-pair library preparation for Illumina sequencing.
Paired-End Sequencing - Acheving maximum coverage across the genome. For example if you have a 300bp contiguous fragment the machine will sequence eg. Mate pair sequencing involves generating long-insert paired-end DNA libraries useful for a number of sequencing applications including.
First PE paired end reads are. Shortinsert pairedend reads SIPERs and long-insert paired-end reads LIPERs. Introduction to Mate Pair Sequencing.
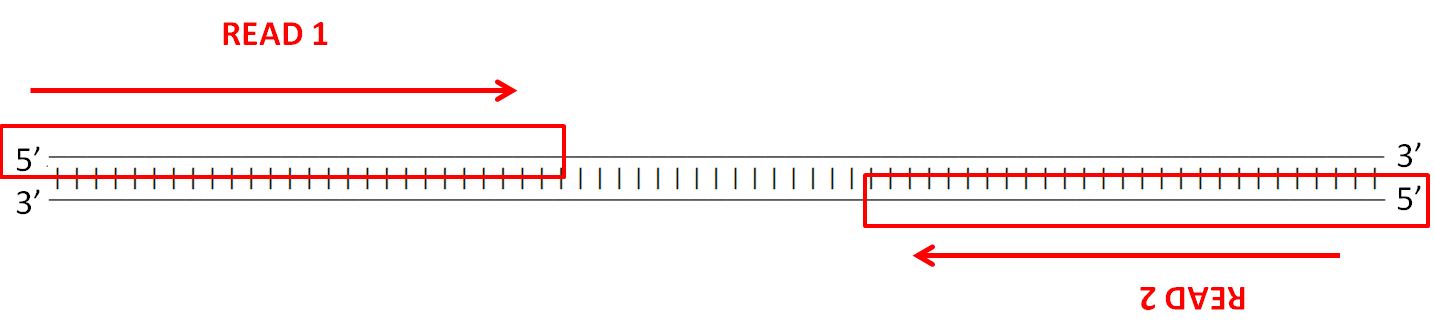
In paired-end sequencing the library preparation yields a set of fragments and the machine sequences each fragment from both ends. Illumina에서 이야기하는 mate pair library는 일종의 jumping library라고 하는 것이 기술적으로 더 정확할 수 있겠다. Paired-end is a type of sequencing.
Paired-end is a type of sequencing. To simplify you can differ between two kinds of reads for paired-end sequencing. Mate-pair is a specific type of library.
In mate-pair sequencing the library preparation yields two. Is Illumina sequencing paired. Paired-end or mate-pair.
The preparation of mate pair libraries is designed to allow classical paired-end sequencing of both ends of a fragment with an original size of several kilobases.

Abi Solid Sequencing Wikipedia
De Novo Genome Assembly Introduction Henrik Lantz Nbissci

Pdf Data Processing Of Nextera Mate Pair Reads On Illumina Sequencing Platforms Semantic Scholar
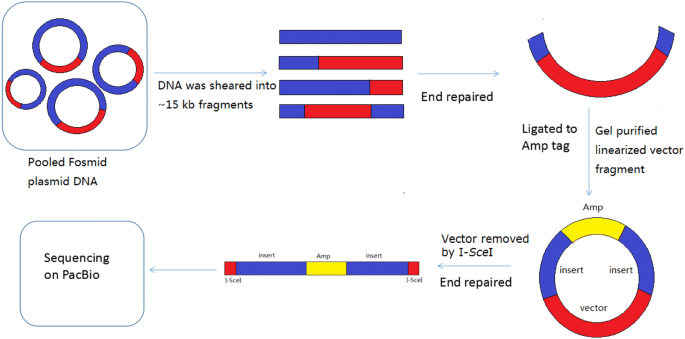
High Throughput Long Paired End Sequencing Of A Fosmid Library By Pacbio Plant Methods Full Text
De Novo Genome Assembly Using Next Generation Sequence
Introduction To Genome Assembly Tutorial

Illumina Sequencing Technology Youtube

Figure 2 From Next Generation Sequencing Platforms Semantic Scholar

Mate Pair Sequencing Paired End Mate Pair Sequencing Paired End Sequencing Youtube

Illumina Sequencing Illumina Sequencing By Synthesis 1010genome Quality Ngs Bioinformatics Data Analysis Services

6 Pairwise Strand Of Paired End And Mate Pair Read Libraries Download Scientific Diagram

Mate Pair Made Possible Enseqlopedia
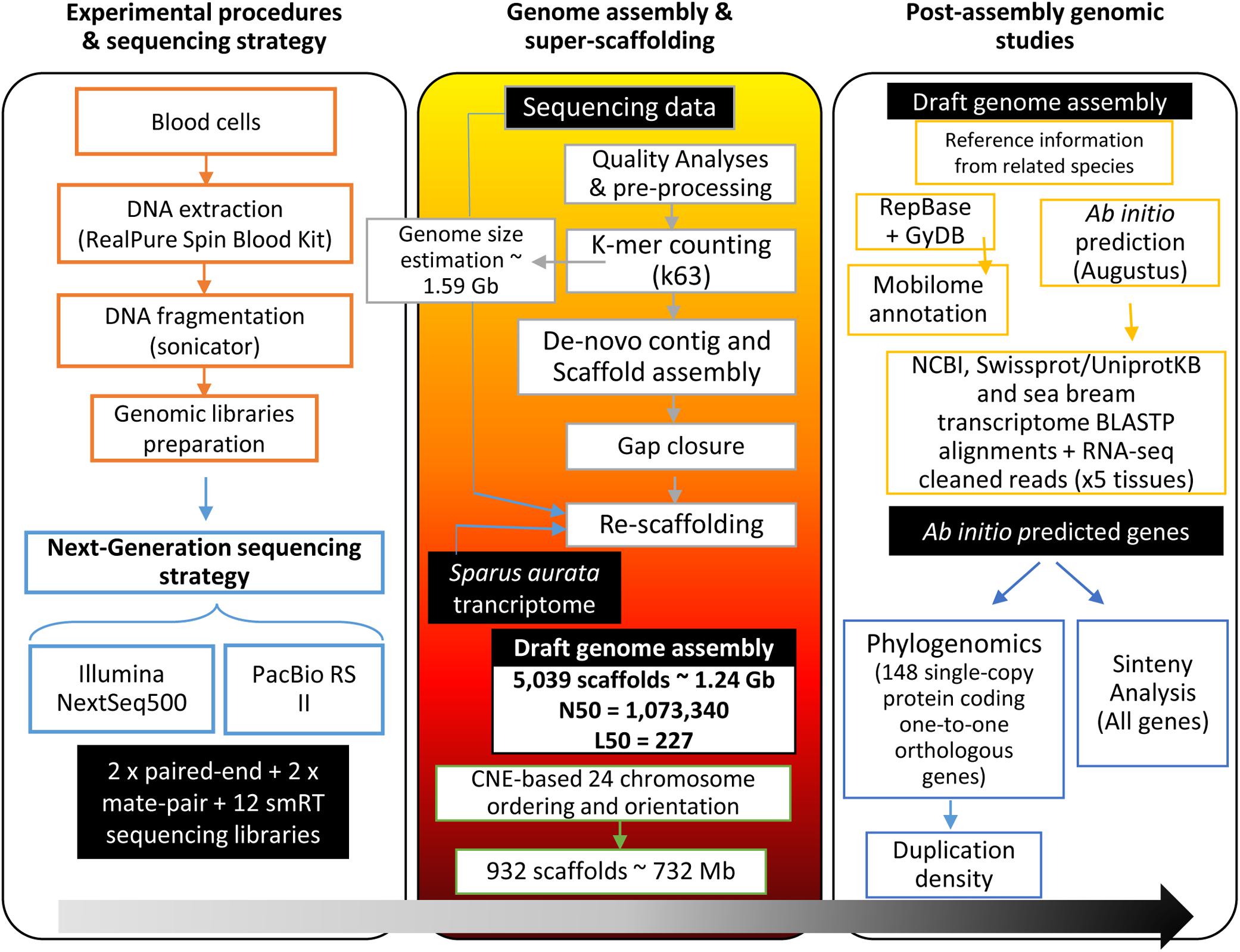
Frontiers Genome Sequencing And Transcriptome Analysis Reveal Recent Species Specific Gene Duplications In The Plastic Gilthead Sea Bream Sparus Aurata
Paired End Sequencing Vs Mate Pair Sequencing Zhongxu Blog
De Novo Genome Assembly Haibao Tang J Craig